
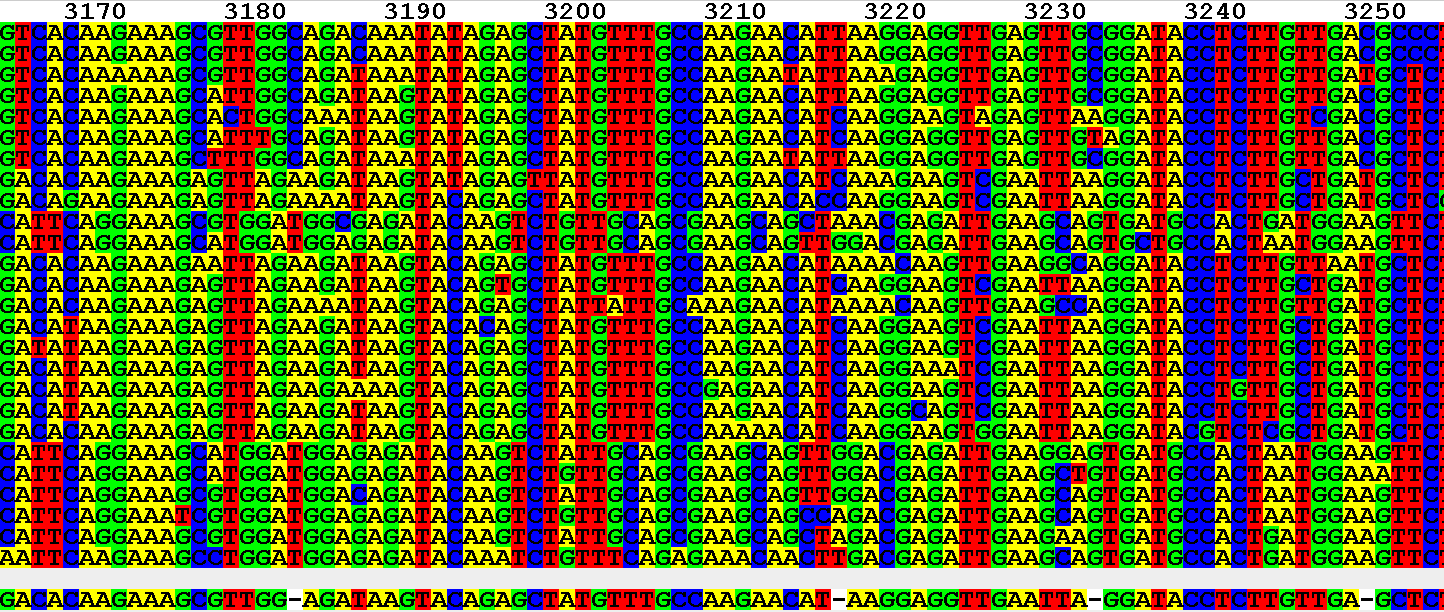
More than 80% of the currently known phylospecies were discovered just within the past 25 years using multilocus Genealogical Concordance Phylogenetic Species Recognition (GCPSR sensu Taylor et al. Published and ongoing molecular phylogenetic analyses indicate this monophyletic genus comprises more than 400 phylogenetically recognizable species distributed among 23 monophyletic lineages referred to as species complexes ( Fig. While the focus of this report is on Fusarium, general aspects of the best practices for DNA sequence-based identification outlined in this report are applicable to other fungi. 2004) and the nonredundant National Center for Biotechnology Information (NCBI) nucleotide collection (i.e., GenBank+EMBL+DDBJ+PDB+RefSeq sequences).

2022 an updated version of FUSARIUM-ID v.1.0 first published by Geiser et al.
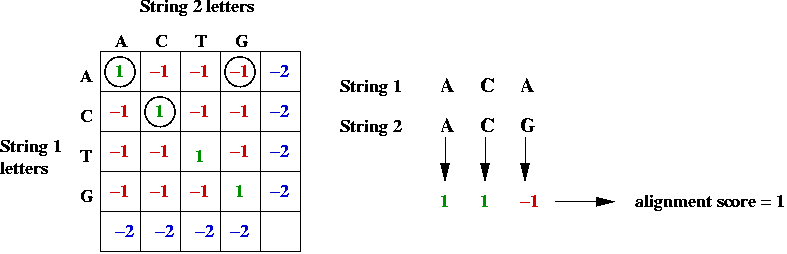
#4peaks align sequence how to#
The primary focus of this special report is to provide instructions on how to identify fusaria by BLASTn analysis of two web-accessible nucleotide databases: FUSARIUM-ID v.3.0 ( Torres-Cruz et al. 2022), are to provide a progress report on our efforts to populate these databases and to outline a set of best practices for DNA sequence-based identification of fusaria.įusarium is a species-rich genus that is arguably the most important group of mycotoxigenic plant pathogens ( Dean et al. The objectives of this Special Report, and its companion in this issue ( Torres-Cruz et al. To address the critical need for accurate pathogen identification, our research groups are focused on populating two web-accessible databases (FUSARIUM-ID v.3.0 and the nonredundant National Center for Biotechnology Information nucleotide collection that includes GenBank) with portions of three phylogenetically informative genes (i.e., TEF1, RPB1, and RPB2) that resolve at or near the species level in every Fusarium species. Paramount among these are: (i) this agriculturally and clinically important genus is currently estimated to comprise more than 400 phylogenetically distinct species (i.e., phylospecies), with more than 80% of these discovered within the past 25 years (ii) approximately one-third of the phylospecies have not been formally described (iii) morphology alone is inadequate to distinguish most of these species from one another and (iv) the current rapid discovery of novel fusaria from pathogen surveys and accompanying impact on the taxonomic landscape is expected to continue well into the foreseeable future. This can be a daunting task for phytopathologists and other applied biologists that need to identify Fusarium in particular, because published and ongoing multilocus molecular systematic studies have highlighted several confounding issues. This is particularly true in publishing peer-reviewed disease reports, where imprecise and/or incorrect identifications weaken the public knowledge base. Accurate species-level identification of an etiological agent is crucial for disease diagnosis and management because knowing the agent’s identity connects it with what is known about its host range, geographic distribution, and toxin production potential.


 0 kommentar(er)
0 kommentar(er)
